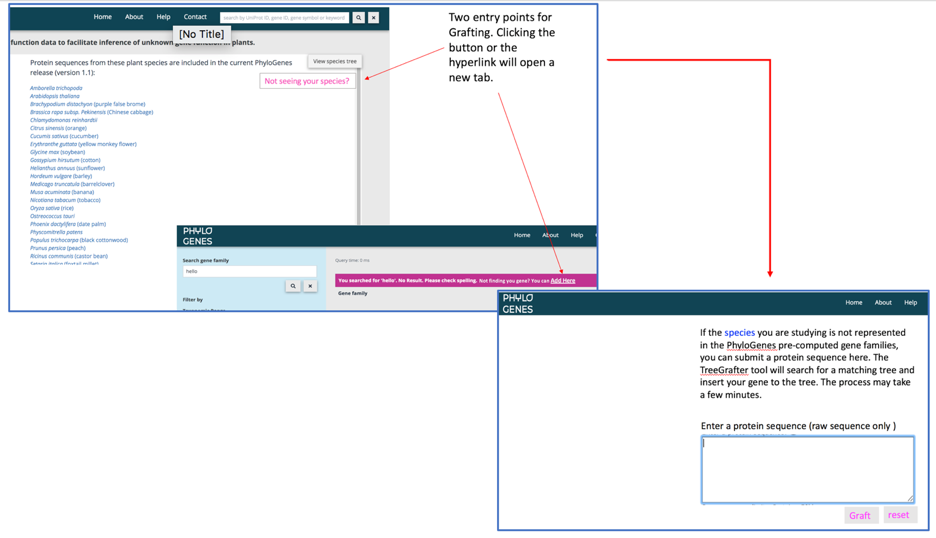
If the species you are studying is not represented in PhyloGenes (check the full species list here), you can submit an individual protein sequence and 'graft' the sequence onto existing precomputed gene families. The TreeGrafter [Tang2019] tool will run HMM scoring and find the best matching gene family, add your sequence to the MSA of that family, then run RAxML to insert your sequence to the best location of the gene family tree. Your sequence will be labeled as 'grafted'.
Note: Grafting works well in many cases that we have tested but there may be instances where unexpected results occur. If this happens to you, please contact us to share the specifics. We are collecting examples so that we can improve the TreeGrafter software.
There are two entry places to take you to the protein sequence input box:
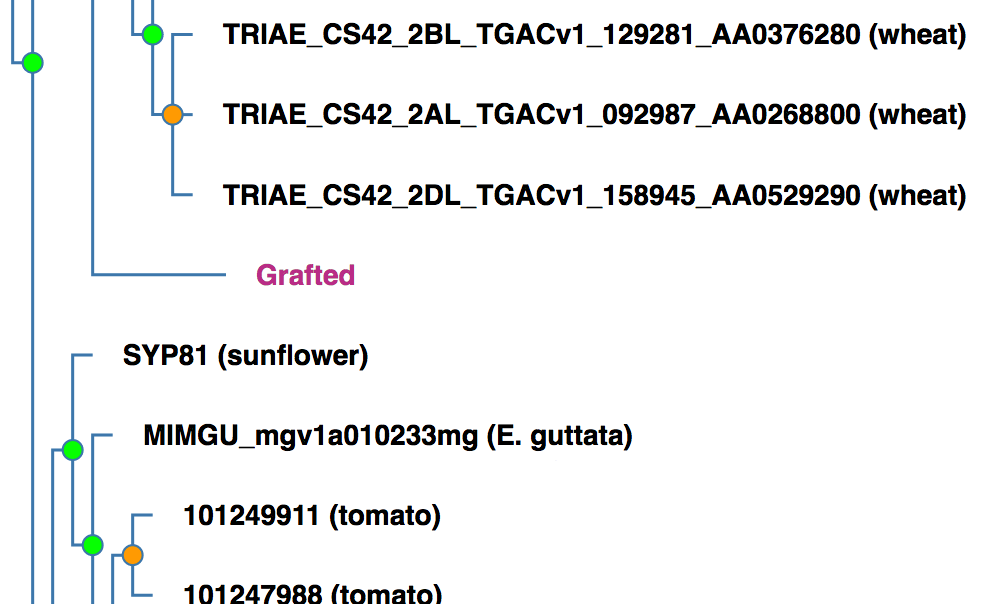
An example of a grafted tree:
Add Comment