Page History
...
Gene families in PhyloGenes are pruned versions of PANTHER gene families (pantherdb.org, ref Mi2019). They contain only genes from selected plant genomes and 10 non-plant model organisms <link to list>(phylogenes.org). Genes from other genomes in the PANTHER build have been removed (pruned) from the PANTHER gene families and gene trees.
...
In PANTHER, gene families are defined as clusters of related protein sequences (each protein sequence represents a distinct gene) for which a good multiple sequence alignment can be made (PubMed:23193289, PubMed:26578592Mi2013, Mi2016). The basic requirements for a family are: (1) the family contains at least five sequences and includes more than one organism, and (2) the family has a sequence alignment of adequate quality to support phylogenetic inference. An alignment must have at least 30 sites aligned across 75% or more of the family members, and the derived Hidden Markov Model (HMM) must be able to recognize, with statistical significance, the sequences used to train it.
...
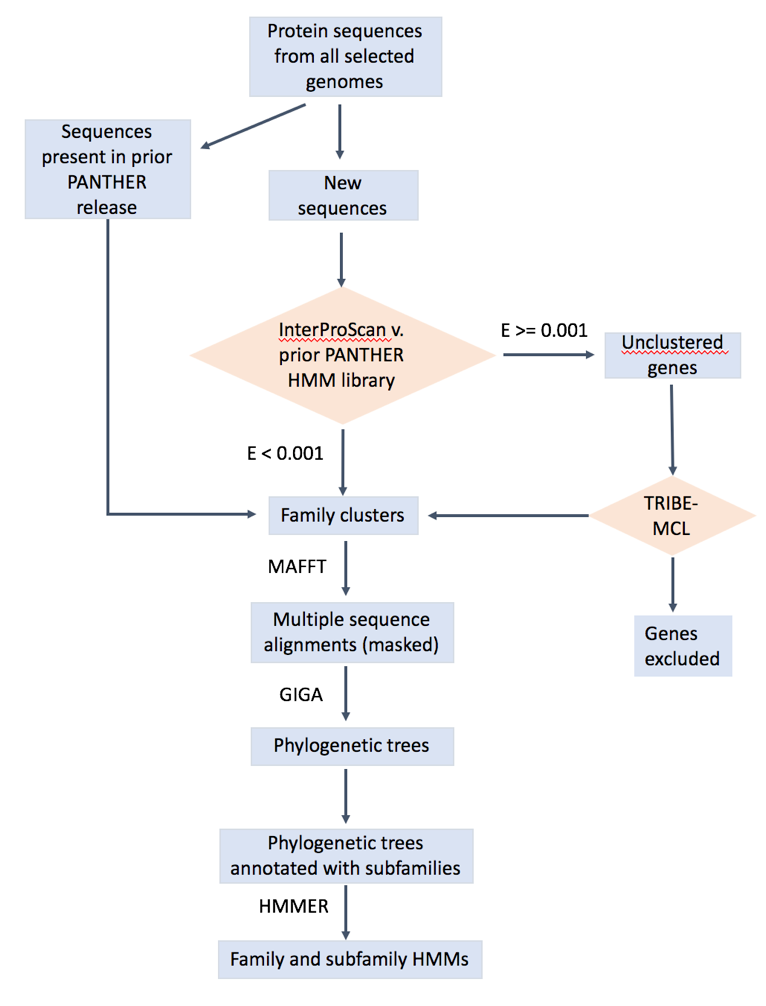
The overall workflow is shown below. The details can be found in : https://www.ncbi.nlm.nih.gov/pubmed/23193289 (https://doi.org/10.1093/nar/gks1118), https://www.ncbi.nlm.nih.gov/pubmed/26578592, https://www.ncbi.nlm.nih.gov/pubmed/27899595 Mi2013, Mi2016, Mi2017 .
How is a PANTHER gene tree constructed?
A PANTHER gene tree is composed of orthologous subtrees (containing protein sequences related by speciation events), joined together by gene duplication (duplication within the same genome) or horizontal transfer (insertion from another genome). PANTHER trees were constructed by using the GIGA algorithm (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-312vThomas2010). The algorithm builds a tree from leaf nodes to the root, using a pairwise sequence distance matrix, a known species tree (based on NCBI taxonomy), and a set of rules to establish the tree topology. Sequence distance is calculated as the fraction of sequence differences between two sequences at selected homologous sites. The homologous sites were selected from multiple sequence alignment of all genes in the family. GIGA iteratively joins together subtrees of sequences, beginning with the two sequences that are closest according to the pairwise sequence distance matrix. The topology of the joined subtree after each iteration is not simply an agglomeration of the constituent subtrees. Rules are used to "rearrange" the joined subtree at each iteration. For example, if a subtree contains only speciation events, the topology is determined by the known species tree. Copying events such as duplication or horizontal transfer are placed within a tree with the most parsimonious solution to minimize gene deletions. The full description of the GIGA algorithm can be found in this paper (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-312Thomas2010).
How is branch length calculated?
First, the ancestral sequence is inferred for each non-leaf node using a local, parsimony-like algorithm that reconstructs each node using only its descendants and closest outgroup. If over half of the descendant nodes align the same amino acid at a given site, it is inferred to be the most likely ancestral amino acid. If the descendants disagree, and the outgroup agrees with one of them, the outgroup amino acid is inferred to be the most likely ancestral amino acid. Otherwise, the ancestral amino acid is considered to be unknown ('X'). Next, the branch length between a parent node and a child node was calculated as the fraction of sequence differences between them. The Jukes-Cantor correction is applied to this value. More details can be found here (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-312Thomas2010).
Why aren’t there any bootstrap or other support values for the tree topology?
...
Subfamilies within each family are groups of genes that share a particularly high degree of protein sequence similarity due to limited divergence from their common ancestor [https://www.ncbi.nlm.nih.gov/pubmed/26578592]. (Mi2016). Subfamilies are, in general, closely-related orthologs. In the PANTHER tree building process, a new subfamily is created within a family after every gene duplication event, or horizontal transfer event. After horizontal transfer, the transferred copy becomes the founder of a new subfamily; the vertically inherited copy remains in the original subfamily. After gene duplication, the copy that changes faster in sequence immediately following the duplication becomes the founder of a new subfamily; the slower-evolving copy remains in the same subfamily. There are two exceptions to this rule: (1) because of the high frequency of gene duplication prior to the vertebrate common ancestor, each vertebrate copy following a gene duplication event founds a new subfamily, and (2) duplicated genes do not found a subfamily if they did not lead to orthologs in at least two extant species.
...
Biologically meaningful family names are assigned by biologist curators (https://www.ncbi.nlm.nih.gov/pubmed/12952881Thomas2003). The curator either assigns a family a more general functional name that applies to all genes in the family (e.g., NUCLEAR HORMONE RECEPTOR) or finds the largest subfamily name (Y) and names the family Y-RELATED. If in the latter case the largest subfamily is “unnamed” then the family is not named.
How are subfamilies named
...
? Why are some subfamilies not named?
The name of a subfamily is transferred from the representative member of the subfamily (https://www.ncbi.nlm.nih.gov/pubmed/26578592Mi2016). If a subfamily contains an annotated SwissProt entry from any of the 12 model organisms (human, mouse, rat, chicken, zebrafish, fruit fly, C. elegans, budding yeast, fission yeast, D. discoideum, Arabidopsis, and E. coli.), then the curated ‘protein name’ is used to name the subfamily. If a subfamily does not contain any SwissProt entries from the model organisms, but contains SwissProt entries from other organisms, the most common SwissProt protein name is used as the subfamily name. If a subfamily does not contain any members from the SwissProt database, a protein name from the TrEMBL entry is automatically selected as the subfamily name. If no name can be found, the subfamily is labeled with ‘unnamed’.
...
Horizontal transfer is the movement of genetic material between organisms other than by the default "vertical" transmission from parent to offspring. Horizontal transfer is common in bacteria. In Eukaryotes, events such as the engulfment of the mitochondrion by the proto-eukaryotic cell will be represented at the level of gene family trees as horizontal transfer from a proteobacterial ancestor to the eukaryotic common ancestor. At each step in PANTHER gene tree building the GIGA algorithm considers the number of gene deletions that would be implied by a history of vertical inheritance, and if that number is too large, a horizontal transfer event is considered to be the more likely interpretation (https://www.ncbi.nlm.nih.gov/pubmed/26578592Mi2016).
Are polyploid organisms represented in gene families?
...
There are two possibilities. It could be that the organism of your gene is not included in the PANTHER pipeline <link to FAQ above>. The list of genomes that are included PANTHER and PhyloGenes can be found here <link>. Alternatively, even if the organism is included in PANTHER and PhyloGenes, not all genes of the genome are part of a gene family. Some genes don’t meet the criteria of being part of a gene family. Statistics covering the current release can be found here <link>.